6 Multiple comparison correction
In whole-brain analysis
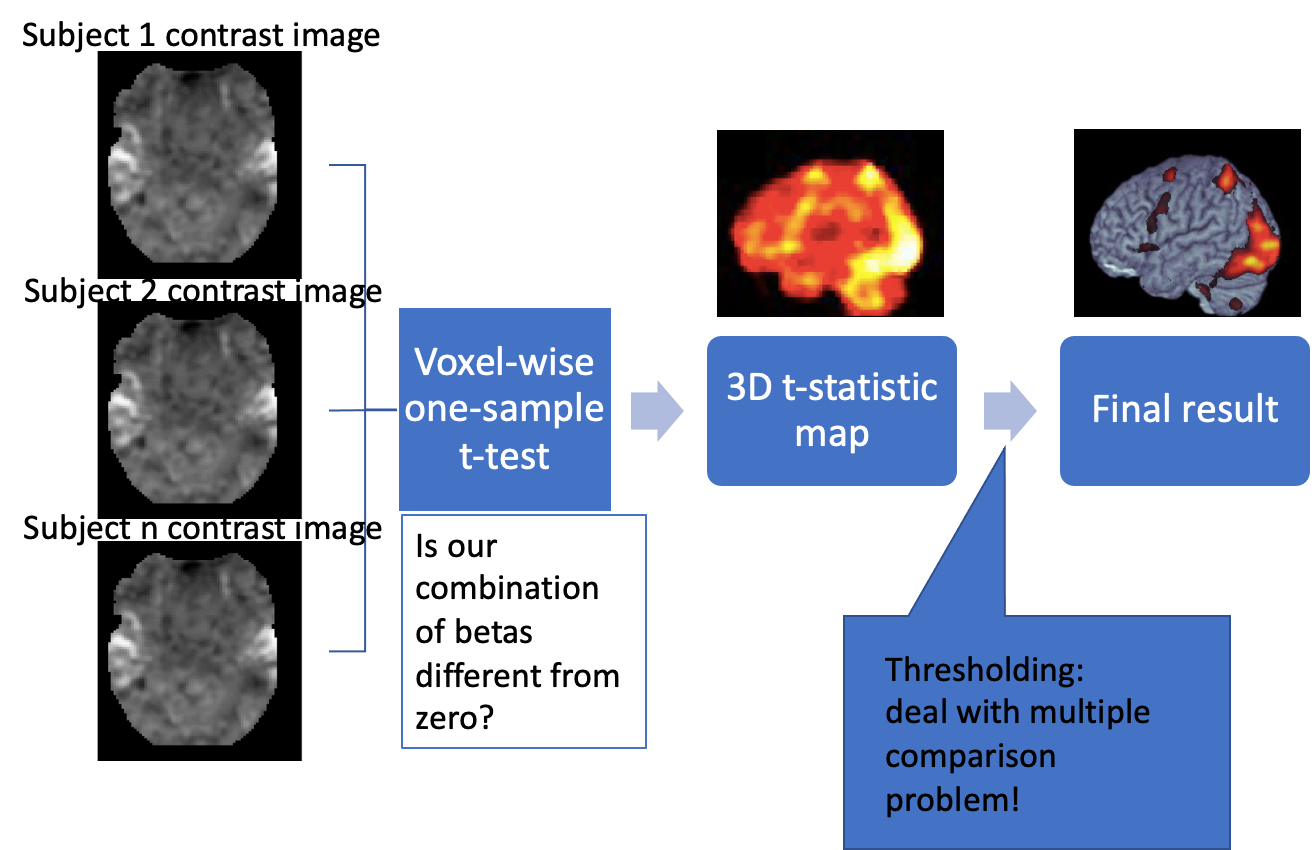
Reminder: group analysis
Family-wise error rate
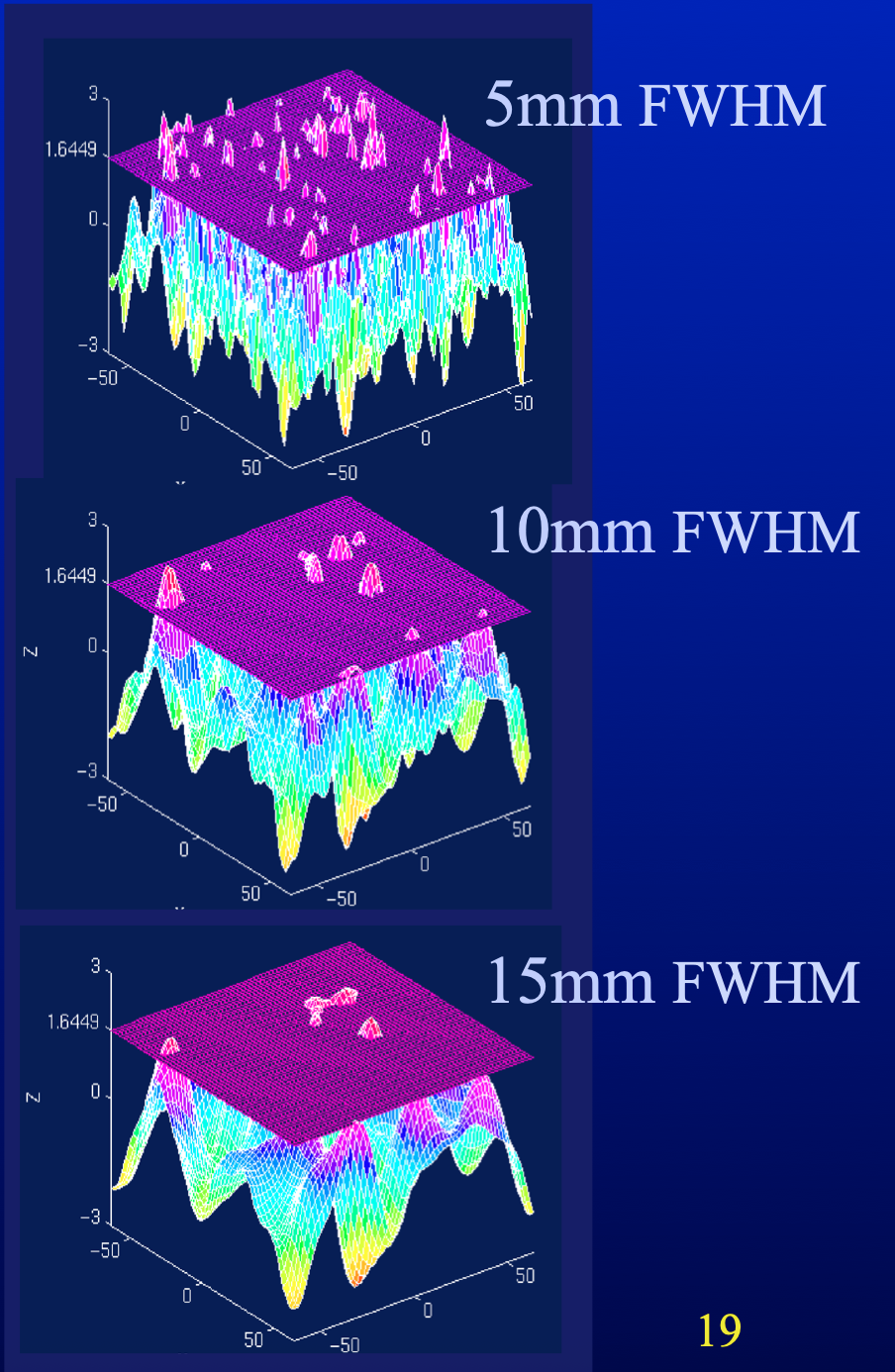
Image credit: unknown
If the data is not smoothed, equivalent to bonferroni
Other correction types
Cluster threshold
False discovery rate
Cluster-level inference
Uncorrected
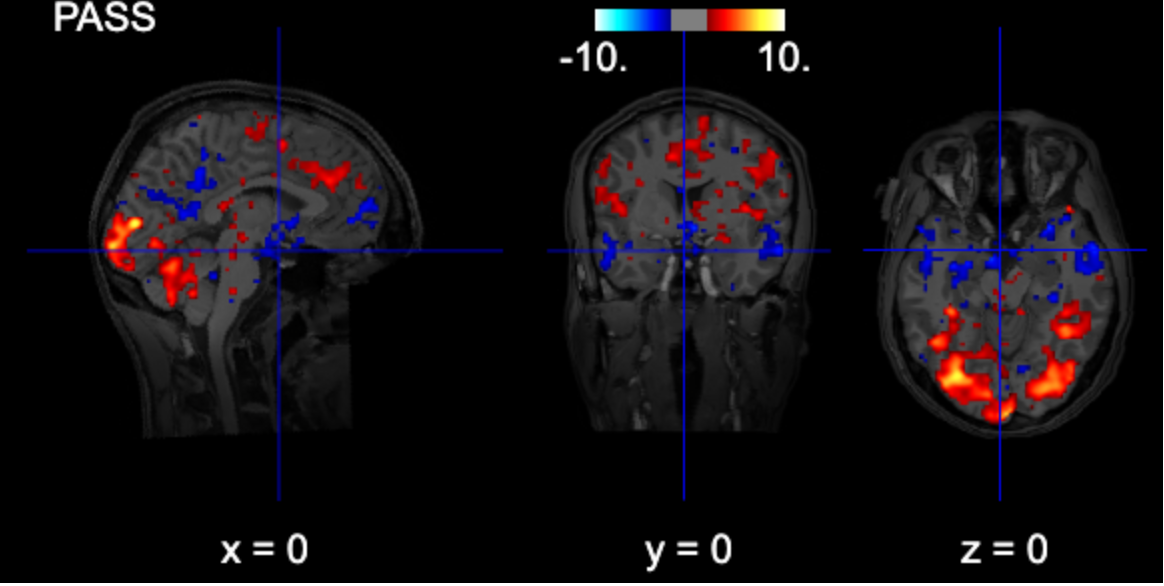
# interactive plot (you can browse the activations)
from nilearn import plotting
# Use subject's anatomy as background
bg_img = '/home/jovyan/gambling/bids/derivatives/fmriprep/sub-001/ses-1/anat/sub-001_ses-1_acq-mprage_desc-preproc_T1w.nii.gz'
plotting.view_img(zmap, threshold=1.96, vmax=10,
bg_img=bg_img,
cut_coords=[0, 0, 0],
width_view=600,
title=contrast_string)Cluster-thresholded
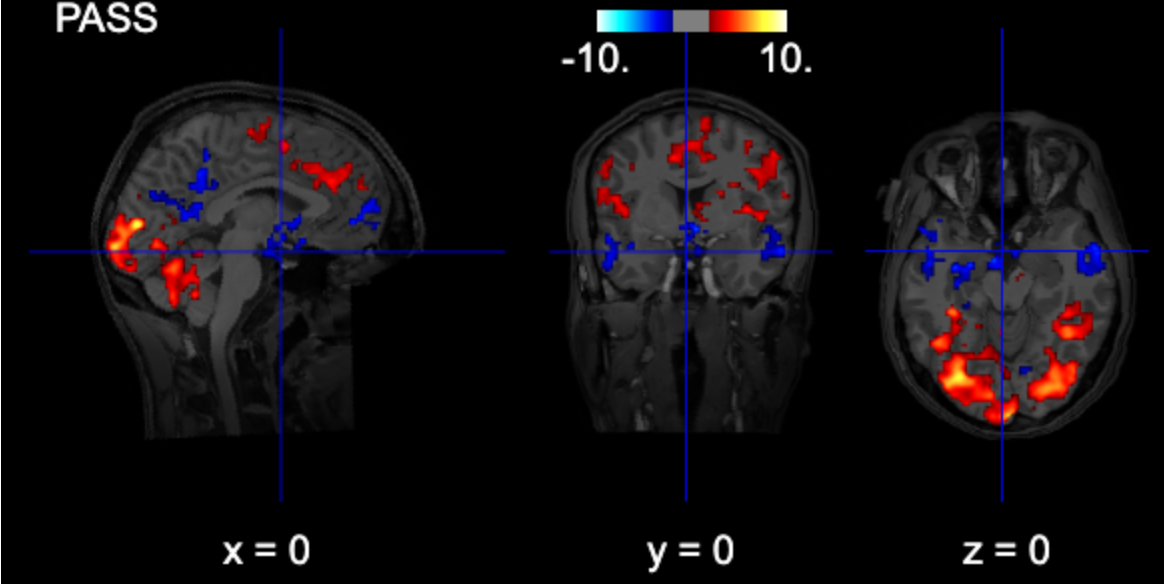
from nilearn.glm import threshold_stats_img
thresholded_map1, threshold1 = threshold_stats_img(
zmap,
alpha=0.05,
height_control="fpr",
cluster_threshold = 100,
two_sided=True,
)
# Use subject's anatomy as background
bg_img = '/home/jovyan/gambling/bids/derivatives/fmriprep/sub-001/ses-1/anat/sub-001_ses-1_acq-mprage_desc-preproc_T1w.nii.gz'
plotting.view_img(thresholded_map1, threshold=threshold1, vmax=10,
bg_img=bg_img,
cut_coords=[0, 0, 0],
width_view=600,
title=contrast_string)FDR-thresholded
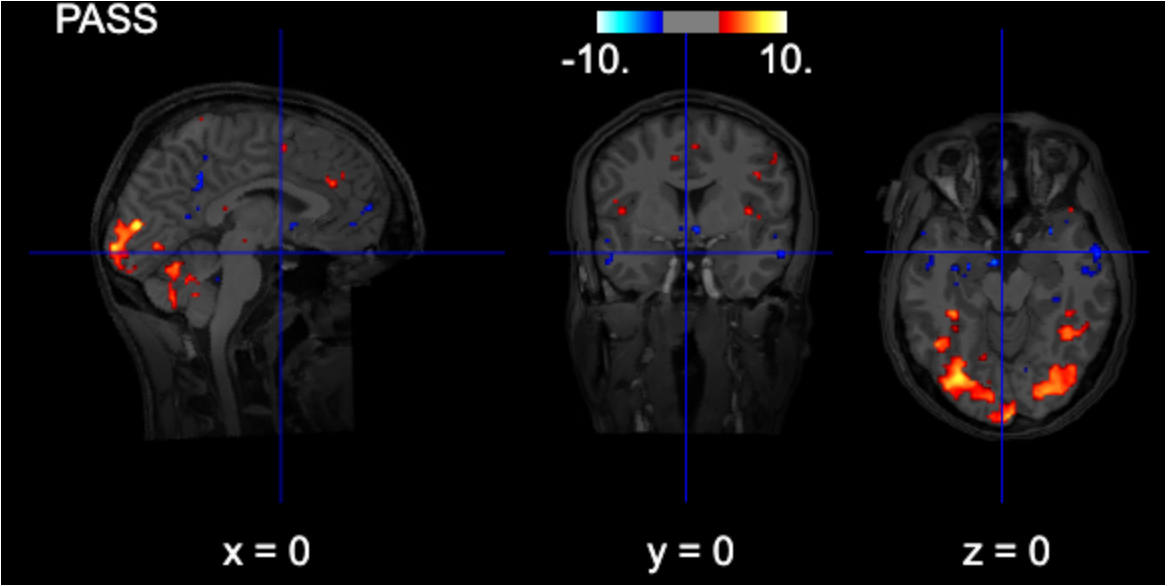
from nilearn.glm import threshold_stats_img
thresholded_map1, threshold1 = threshold_stats_img(
zmap,
alpha=0.05,
height_control="fdr",
two_sided=True,
)
# Use subject's anatomy as background
bg_img = '/home/jovyan/gambling/bids/derivatives/fmriprep/sub-001/ses-1/anat/sub-001_ses-1_acq-mprage_desc-preproc_T1w.nii.gz'
plotting.view_img(thresholded_map1, threshold=threshold1, vmax=10,
bg_img=bg_img,
cut_coords=[0, 0, 0],
width_view=600,
title=contrast_string)FWE-thresholded
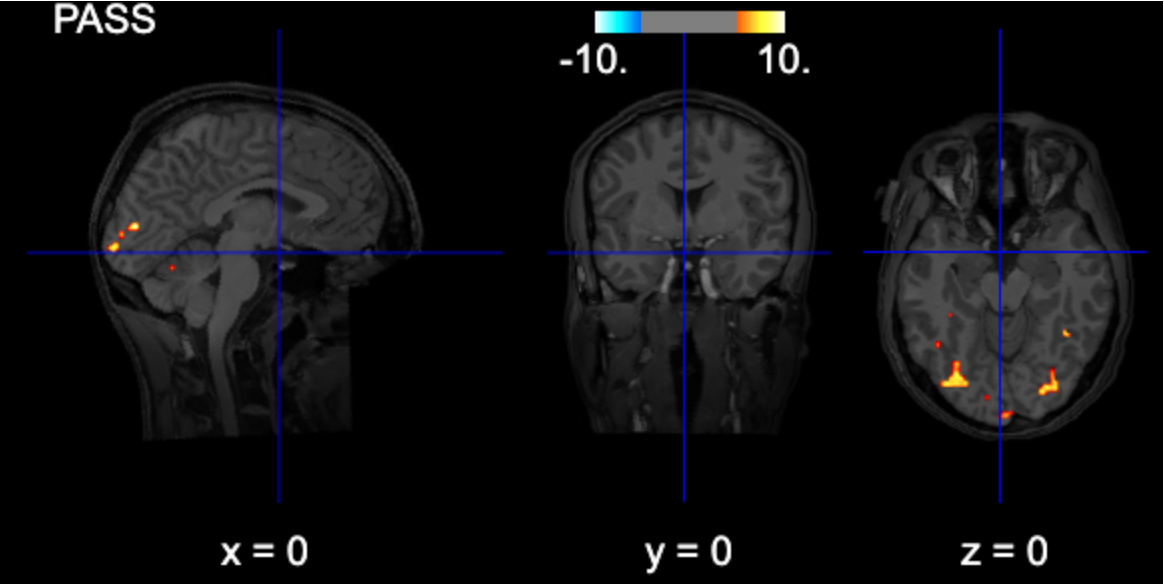
from nilearn.glm import threshold_stats_img
thresholded_map1, threshold1 = threshold_stats_img(
zmap,
alpha=0.05,
height_control="bonferroni",
two_sided=True,
)
# Use subject's anatomy as background
bg_img = '/home/jovyan/gambling/bids/derivatives/fmriprep/sub-001/ses-1/anat/sub-001_ses-1_acq-mprage_desc-preproc_T1w.nii.gz'
plotting.view_img(thresholded_map1, threshold=threshold1, vmax=10,
bg_img=bg_img,
cut_coords=[0, 0, 0],
width_view=600,
title=contrast_string)Non-parametric inference
Permutation-based inference

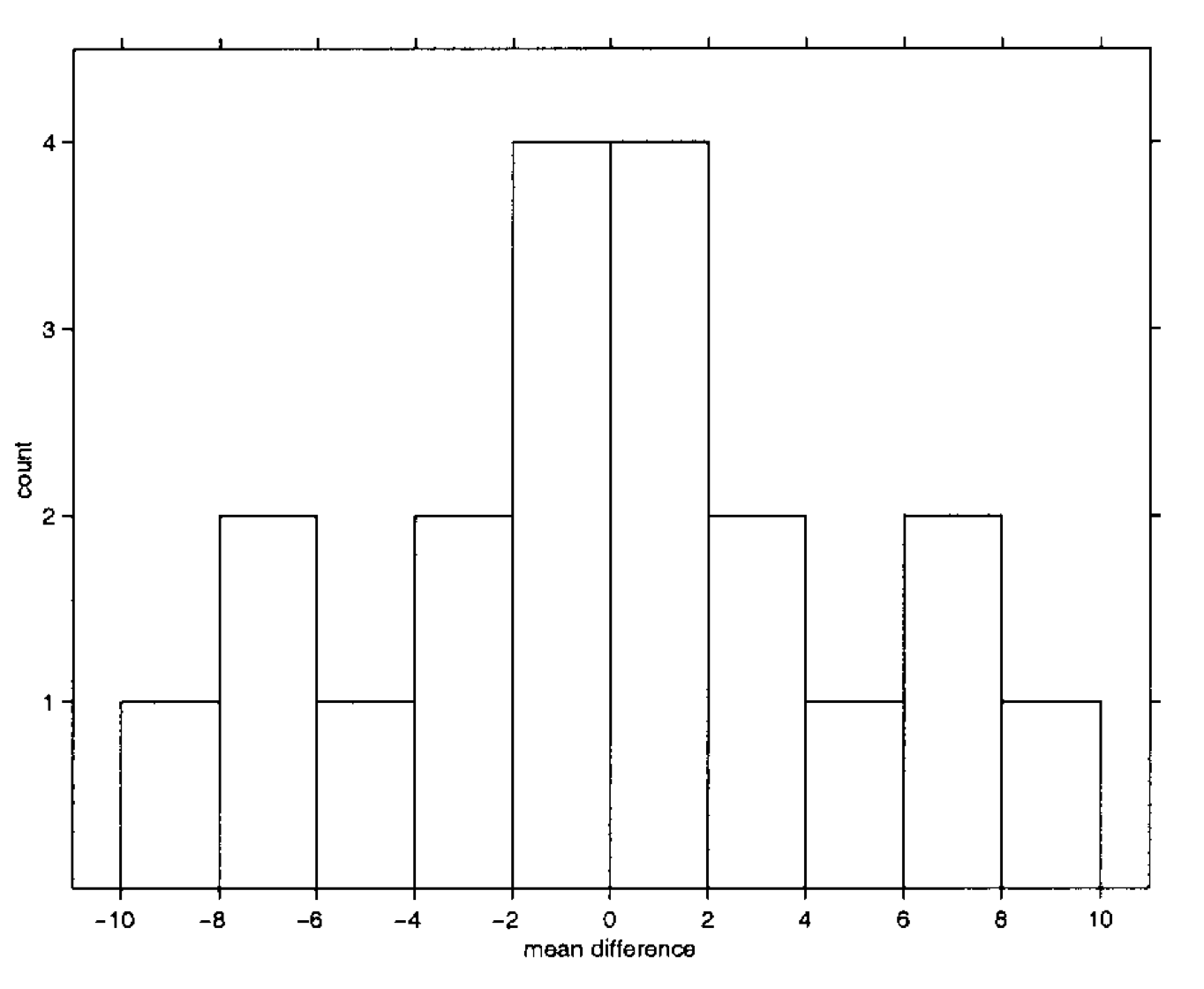
For the multiple comparison case, we use the same logic, but instead of single voxel we consider the maximum of each statistical map.
References
Nichols, Thomas E., and Andrew P. Holmes. 2001. “Nonparametric Permutation Tests for Functional Neuroimaging: A Primer with Examples.” Human Brain Mapping 15 (1): 1–25. https://doi.org/10.1002/hbm.1058.
